TxTrack and GroupFeatureTrack
Source:vignettes/examples/tracktype-TxTrack.Rmd
tracktype-TxTrack.RmdA TxTrack displays transcripts and their stucture (exons, coding sequence) along genomic coordinate. It can be constructed from either a TxDb object by TxTrackFromTxDb or a GRanges object by TxTrackFromGRanges.
For the constructing method from GRanges, two necessary meta-columns (“type”, “tx_id”) of the GRanges are required. The “tx_id” indicates grouping, and the “type” can be “exon” or “cds” by which ranges of “cds” will be filled with color. This method can be used together with biovizBase::crunch to fetch gene model in a certain region or of a certain gene from TxDb or EnsDb.
The constructed track may be further modified to adjust color, tooltip, display labels, etc.
Similar to the relationship between GeneTrack and FeatureTrack, when the display method of TxTrack applied to genomic feature that may have gaps on genomic coordinate, e.g. RNA-related features that are mapped to genomic coordinate, it is called GroupFeatureTrack. It can be constructed from a GRangesList object by GroupFeatureTrack function, assuming ranges in each group are on the same strand and do not overlap.
suppressPackageStartupMessages({
library(TnT)
library(GenomicFeatures)
library(TxDb.Hsapiens.UCSC.hg19.knownGene)
library(EnsDb.Hsapiens.v75)
})
txdb <- TxDb.Hsapiens.UCSC.hg19.knownGeneTranscript Track From TxDb
txtrack <- TnT::TxTrackFromTxDb(txdb, height = 400)
TnTGenome(txtrack, view.range = GRanges("chr17", IRanges(41190000, 41290000)))## Warning in readLines(con): incomplete final line found on '/home/jialin/R/
## x86_64-suse-linux-gnu-library/3.4/TnT/htmlwidgets/trackWidget.yaml'Please note that TxTrackFromTxDb currently consumes a relative large amount of memory, currently you can use seqlevel argument to limit the chromosomes to extract data from, e.g. TxTrackFromTxDb(txdb, seqlevel = "chr17"). But in future, we will use S4 class CompressedSplitDataFrameList to store the exons and implement direct conversion method to JSON to avoid the intermediate structure as list of data frame.
Transcript Track From GRanges (Fetched From EnsDb)
We can use function crunch from biovizBase package to fetch gene model within a certain genomic region or of a certain gene.
gr <- biovizBase::crunch(EnsDb.Hsapiens.v75, ~ symbol == "BRCA2")## Fetching data...OK
## Parsing exons...OK
## Defining introns...OK
## Defining UTRs...OK
## Defining CDS...OK
## aggregating...
## Donegr## GRanges object with 230 ranges and 4 metadata columns:
## seqnames ranges strand | tx_id
## <Rle> <IRanges> <Rle> | <character>
## ENSE00001184784 13 [32889611, 32889804] + | ENST00000380152
## ENSE00002209788 13 [32889617, 32889804] + | ENST00000544455
## ENSE00002143308 13 [32889642, 32889804] + | ENST00000530893
## ENSE00001484009 13 [32890559, 32890664] + | ENST00000380152
## ENSE00003339705 13 [32890559, 32890660] + | ENST00000530893
## ... ... ... ... . ...
## ENST00000544455 13 [32953887, 32954050] + | ENST00000544455
## ENST00000544455 13 [32954144, 32954282] + | ENST00000544455
## ENST00000544455 13 [32968826, 32969070] + | ENST00000544455
## ENST00000544455 13 [32971035, 32971181] + | ENST00000544455
## ENST00000544455 13 [32972299, 32972907] + | ENST00000544455
## gene_name gene_id type
## <character> <character> <factor>
## ENSE00001184784 BRCA2 ENSG00000139618 exon
## ENSE00002209788 BRCA2 ENSG00000139618 exon
## ENSE00002143308 BRCA2 ENSG00000139618 exon
## ENSE00001484009 BRCA2 ENSG00000139618 exon
## ENSE00003339705 BRCA2 ENSG00000139618 exon
## ... ... ... ...
## ENST00000544455 BRCA2 ENSG00000139618 cds
## ENST00000544455 BRCA2 ENSG00000139618 cds
## ENST00000544455 BRCA2 ENSG00000139618 cds
## ENST00000544455 BRCA2 ENSG00000139618 cds
## ENST00000544455 BRCA2 ENSG00000139618 cds
## -------
## seqinfo: 1 sequence from GRCh37 genomeThen we use this GRanges object to construct the TxTrack.
brca2tx <- TnT::TxTrackFromGRanges(gr, label = "BRCA2, transcripts", color = "grey2", height = 300)
TnTGenome(brca2tx)## - Missing argument `view.range`:
## automatically select 32879087..32984329 on seqlevel 13...## Warning in readLines(con): incomplete final line found on '/home/jialin/R/
## x86_64-suse-linux-gnu-library/3.4/TnT/htmlwidgets/trackWidget.yaml'Let’s compare it with output of ggbio!
library(ggbio)## Loading required package: ggplot2## No methods found in "RSQLite" for requests: dbGetQuery## Need specific help about ggbio? try mailing
## the maintainer or visit http://tengfei.github.com/ggbio/##
## Attaching package: 'ggbio'## The following objects are masked from 'package:ggplot2':
##
## geom_bar, geom_rect, geom_segment, ggsave, stat_bin,
## stat_identity, xlimggplot() + geom_alignment(EnsDb.Hsapiens.v75, which = ~ symbol == "BRCA2")## Fetching data...## OK## Parsing exons...OK
## Defining introns...OK
## Defining UTRs...OK
## Defining CDS...OK
## aggregating...
## Done
## Constructing graphics...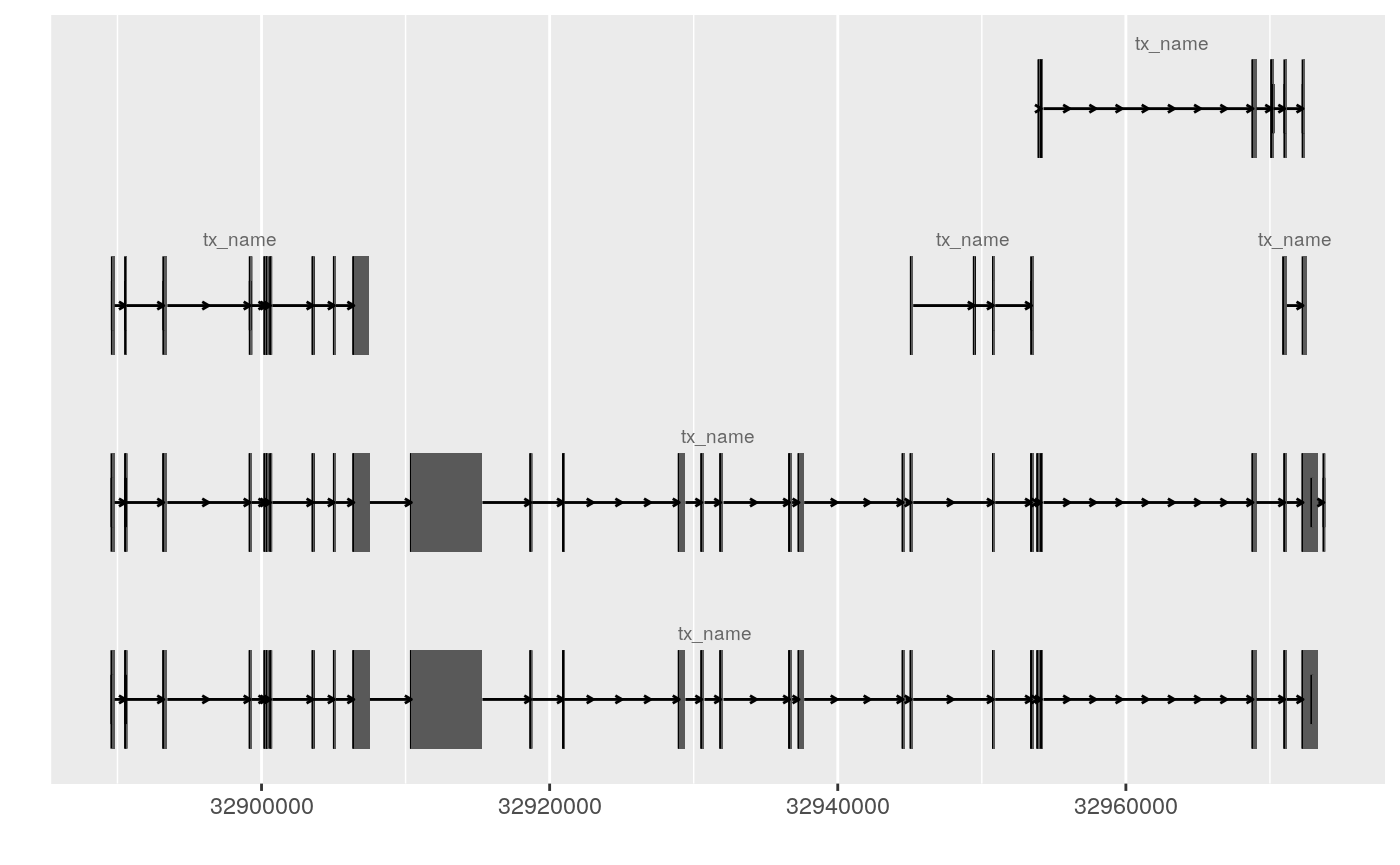
Modify Track Color and Tooltip
Following the example above, we can fill the tooltip with more information and change the colors.
brca2tx$tooltip <- select(EnsDb.Hsapiens.v75,
keys = brca2tx$tooltip$tx_id, keytype = "TXID", columns = c("GENEID", "SYMBOL", "TXBIOTYPE")
)
brca2tx$color <- TnT::mapcol(brca2tx$tooltip$TXBIOTYPE)
brca2tx$display_label <- TnT::strandlabel(
paste(brca2tx$tooltip$SYMBOL, brca2tx$tooltip$TXBIOTYPE), strand(TnT::trackData(brca2tx))
)
TnTGenome(brca2tx)## - Missing argument `view.range`:
## automatically select 32879087..32984329 on seqlevel 13...## Warning in readLines(con): incomplete final line found on '/home/jialin/R/
## x86_64-suse-linux-gnu-library/3.4/TnT/htmlwidgets/trackWidget.yaml'GroupFeatureTrack From GRangesList
grl <- GRangesList(
GRanges("chr12", IRanges(c(100, 200, 1000), width = 50), strand = "+"),
GRanges("chr12", IRanges(c(900, 1300, 1400), width = 50), strand = "-"),
GRanges("chr12", IRanges(c(900, 1300, 1400), width = 50), strand = "-"),
GRanges("chr12", IRanges(c(900, 1300, 1400), width = 50), strand = "*"),
GRanges("chr12", IRanges(c(900, 1300, 1400), width = 50), strand = "-"),
GRanges("chr12", IRanges(c(900, 1300, 1400), width = 50), strand = "-"),
GRanges()
)
names(grl) <- LETTERS[1:7]
grltrack <- TnT::GroupFeatureTrack(grl, color = "steelblue")
TnTGenome(grltrack)## - Missing argument `view.range`:
## automatically select -69..1617 on seqlevel chr12...## - Missing argument `coord.range` and seqlength is unknown:
## automatically set coordinate limit to -189..1738 ...## Warning in readLines(con): incomplete final line found on '/home/jialin/R/
## x86_64-suse-linux-gnu-library/3.4/TnT/htmlwidgets/trackWidget.yaml'Session Info
sessionInfo()## R version 3.4.1 (2017-06-30)
## Platform: x86_64-suse-linux-gnu (64-bit)
## Running under: openSUSE Tumbleweed
##
## Matrix products: default
## BLAS: /usr/lib64/R/lib/libRblas.so
## LAPACK: /usr/lib64/R/lib/libRlapack.so
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] parallel stats4 methods stats graphics grDevices utils
## [8] datasets base
##
## other attached packages:
## [1] ggbio_1.26.1
## [2] ggplot2_2.2.1
## [3] EnsDb.Hsapiens.v75_2.99.0
## [4] ensembldb_2.1.13
## [5] AnnotationFilter_1.1.9
## [6] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
## [7] GenomicFeatures_1.30.3
## [8] AnnotationDbi_1.40.0
## [9] Biobase_2.38.0
## [10] TnT_1.3.1
## [11] GenomicRanges_1.30.3
## [12] GenomeInfoDb_1.14.0
## [13] IRanges_2.12.0
## [14] S4Vectors_0.16.0
## [15] BiocGenerics_0.24.0
##
## loaded via a namespace (and not attached):
## [1] colorspace_1.3-2 rprojroot_1.3-2
## [3] biovizBase_1.25.1 htmlTable_1.11.1
## [5] XVector_0.18.0 base64enc_0.1-3
## [7] fs_1.2.2 dichromat_2.0-0
## [9] rstudioapi_0.7 roxygen2_6.0.1
## [11] bit64_0.9-7 interactiveDisplayBase_1.15.0
## [13] xml2_1.1.1 splines_3.4.1
## [15] knitr_1.20 Formula_1.2-2
## [17] jsonlite_1.5 Rsamtools_1.30.0
## [19] cluster_2.0.6 graph_1.55.0
## [21] shiny_1.0.5 compiler_3.4.1
## [23] httr_1.3.1 backports_1.1.2-9000
## [25] assertthat_0.2.0 Matrix_1.2-10
## [27] lazyeval_0.2.1 acepack_1.4.1
## [29] htmltools_0.3.6 prettyunits_1.0.2
## [31] tools_3.4.1 gtable_0.2.0
## [33] GenomeInfoDbData_1.0.0 reshape2_1.4.3
## [35] Rcpp_0.12.16 pkgdown_1.0.0
## [37] Biostrings_2.46.0 rtracklayer_1.38.3
## [39] stringr_1.3.0 mime_0.5
## [41] XML_3.98-1.10 AnnotationHub_2.9.17
## [43] zlibbioc_1.24.0 MASS_7.3-47
## [45] scales_0.5.0.9000 BSgenome_1.46.0
## [47] VariantAnnotation_1.24.5 BiocInstaller_1.28.0
## [49] ProtGenerics_1.9.1 SummarizedExperiment_1.8.1
## [51] RBGL_1.53.0 RColorBrewer_1.1-2
## [53] yaml_2.1.19 curl_3.2
## [55] memoise_1.1.0 gridExtra_2.3
## [57] biomaRt_2.34.2 rpart_4.1-11
## [59] reshape_0.8.7 latticeExtra_0.6-28
## [61] stringi_1.2.2 RSQLite_2.1.0
## [63] desc_1.2.0 RMySQL_0.10.14
## [65] checkmate_1.8.5 BiocParallel_1.12.0
## [67] rlang_0.2.0.9001 pkgconfig_2.0.1
## [69] commonmark_1.4 matrixStats_0.52.2
## [71] bitops_1.0-6 evaluate_0.10.1
## [73] lattice_0.20-35 labeling_0.3
## [75] GenomicAlignments_1.14.2 htmlwidgets_1.2
## [77] bit_1.1-12 GGally_1.3.2
## [79] plyr_1.8.4 magrittr_1.5
## [81] R6_2.2.2 Hmisc_4.1-1
## [83] DelayedArray_0.4.1 DBI_0.8
## [85] pillar_1.2.1 foreign_0.8-69
## [87] survival_2.41-3 RCurl_1.95-4.10
## [89] nnet_7.3-12 tibble_1.4.2
## [91] crayon_1.3.4 OrganismDbi_1.19.0
## [93] rmarkdown_1.9 progress_1.1.2
## [95] grid_3.4.1 data.table_1.11.0
## [97] blob_1.1.1 digest_0.6.15
## [99] xtable_1.8-2 httpuv_1.3.6.2
## [101] munsell_0.4.3